Targeted Next-generation Sequencing for Reliable Detection of Genetic Status in Breast Cancer
By Hassan Tariq1, Asma Gul1, Muhammad Zubair1, Syed Raza Jaffer2, Nadeem Zafar1, Ghazala Sadaf1Affiliations
doi: 10.29271/jcpsp.2021.07.837ABSTRACT
Objective: To test the germline oncogenic mutations in BRCA1 and BRCA2 genes, associated with triple-negative breast cancer (TNBC) patients under study, by targeted sequencing of their DNA with next-generation sequencing (NGS) platform.
Study Design: Cross-sectional observational study.
Place and Duration of Study: Histopathology Department, Armed Forces Institute of Pathology (AFIP) Rawalpindi, Pakistan from May to June 2020.
Methodology: Peripheral blood of 14 women (aged ≤60) with triple negative breast carcinoma (TNBC) was taken with the consent of performing germline genetic testing. Targeted NGS was performed for all coding regions and splicing sites of BRCA1 and BRCA2 genes, using AmpliSeq for Illumina BRCA Panel and Illumina MiSeq sequencer (placed at AFIP). Analysis and interpretation of the sequencing results have been done by using Illumina bioinformatics tools and external databases.
Results: Two hundred and fifty-four variants were detected in BRCA1 and BRCA2 genes, having variant quality score of 100 in all cases under study. As a result, two pathogenic variants and three variants of uncertain significance were interpreted in this germline pipeline. Cases with pathogenic variants had early onset breast cancer with age less than 35.
Conclusion: Germline variants in BRCA were detected in the known cases of TNBC, which will not only identify the most prevalent mutations in this region; but will also make them a candidate to receive targeted therapies, which was previously not possible without genetic testing. Moreover, this study further validates the importance of early BRCA genetic screening in young patients, who have positive family history of breast carcinoma.
Key Words: Breast cancer, Triple negative, Next-generation sequencing, BRCA1, BRCA2.
INTRODUCTION
Breast cancer is the most prevalent type in the world of all cancers, and in Pakistani women, too. In Pakistan, 30.8% deaths of cancer suffering women are by breast cancer.1 Triple-negative breast cancer (TNBC) accounts for about 20% of all cases of breast cancer, it is an aggressive subtype of breast carcinoma with poor prognosis and a higher risk of recurrence.2 TNBC is defined by the lack of expression of human epidermal growth factor receptor two (HER2/neu), estrogen receptor (ER) and progesterone receptors (PR). BRCA1 and BRCA2 are tumor-suppressor genes and involved in DNA damage repair and recombination, cell-cycle checkpoint control, apoptosis and transcriptional regulation.3 Variants in BRCA genes induce defective DNA repair mechanisms, which are associated with hereditary breast and/or ovarian cancers.4
According to NCCN guidelines, breast carcinoma patients with triple negative immunohistochemical markers, estrogen, progesterone and HER2/neu, with age equal or less than 60, should be screened for BRCA mutations/variants.5 Previous explorations have validated that the cancer patients, who are BRCA-mutated, are sensitive to platinum chemotherapy and show a good prognosis after treatment with poly ADP ribose polymerase (PARP) inhibitors (PARPi).6,7 Food and Drug Administration (FDA) has approved the PARPi that target and kill HER2/neu negative BRCA mutated cancer cells.8 Gene BRCA1 is located on chromosome 17, and contains 24 exons that code for 1863 amino acids (NM_00,724.3), whereas gene BRCA2 is located on chromosome 13, and contains 27 exons that code for 3418 amino acids (NM_000059.3).9,10 BRCA1/2 genes are highly polymorphic regions and there are no evident mutation hot spots, except in the Ashkenazi population.11,12
Literature show that germline BRCA mutations are highly penetrant in breast or ovarian cancer, which mostly run as autosomal dominant form.13 BRCA1 mutations are more likely to cause TNBC than BRCA2 mutations.14 In a comprehensive study of genetic testing of 192 TNBC cases in Pakistan, Sanger sequencing method was used for screening 26 hotspot mutations in BRCA1 and BRCA2. One hundred and twenty-five cases with pathogenic mutations were identified, in which 84% variants are of BRCA1 and 16% of BRCA2 detected.15
Next-generation sequencing (NGS) system is going to establish for genetic testing of breast cancer. The aim of this study was to test germline variants associated with TNBC cases, by sequencing all exons and splice site regions of BRCA1 and BRCA2 genes via NGS.
METHODOLOGY
It was an observational study where the participants were recruited from the records of Histopathology Department of AFIP, Rawalpindi, Pakistan, diagnosed as triple negative breast cancers by doing immunohistochemistry for ER, PR and HER2/neu receptors and were scored quantitatively following College of American Pathologists (CAP) guidelines16,17 with simultaneous positive internal control. The inclusion criteria was: females having triple negative breast carcinoma, aged equal or less than 60 years. Exclusion criteria: were triple-negative breast cancer cases aged more than 60 years. Fourteen cases were enrolled for this study with informed consent. This project was approved by the Ethical Committee of the Armed Forces Institute of Pathology (AFIP), Pakistan.
Peripheral blood samples in EDTA vials were collected at AFIP in May 2020. Genomic DNA isolation from the whole blood for germline study of breast cancer, was performed using QIAamp Blood DNA Mini Kit (Qiagen, Hilden, Germany) and quantified using Qubit assay according to manufacturer’s instructions (Thermo Fisher Scientific, USA). Isolated genomic DNA was diluted to the required concentration for polymerase chain reaction (PCR)-based library preparation. Targeted amplification of the coding regions and splicing sites of BRCA1 (NM 007294) and BRCA2 (NM 000059) genes was performed using AmpliSeq for Illumina BRCA Panel. The uniquely indexed BRCA panel libraries were prepared according to the AmpliSeq for Illumina BRCA Panel Reference Guide and were proceeded to paired-end sequencing by synthesis utilising MiSeq sequencer (Illumina, San Diego, CA, USA). Local run manager of MiSeq sequencer analysed the base quality and amplicon coverage of raw reads. Then the cleaned reads, which passed the quality phred score more than 30, were aligned to the human reference genome hg19/GRCh37 via BWA-MEM Whole-Genome (Aligner) version 0.7.9a-isis-1.0.2 and subsequently called mismatched calls as variants by Pisces Variant Caller version 5.2.9.23. Genetic variants were identified and annotated via Illumina Annotation Engine version 2.0.11-0-g7fb24a09.
Interpretation of variants was done according to the guidelines of American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP).18 To categorise the variants into pathogenic, likely pathogenic, variant of uncertain significance (VUS), likely benign or benign; in-silico prediction software and external curated databases were used. On the basis of the in-silico prediction output, SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and Mutation Taster (http://www.mutationtaster.org/), functional effect of resultant variants on the protein of BRCA1 and BRCA2 genes were identified. For minor allele frequencies of resultant variants, genome aggregation database (https://gnomad. broadinstitute.org/) was used. ClinVar (https://www.ncbi.nlm. nih.gov/clinvar/), dbSNP (https://www.ncbi.nlm.nih.gov/snp/) and Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/) databases were consulted for the respective variants to examine their clinical significance.
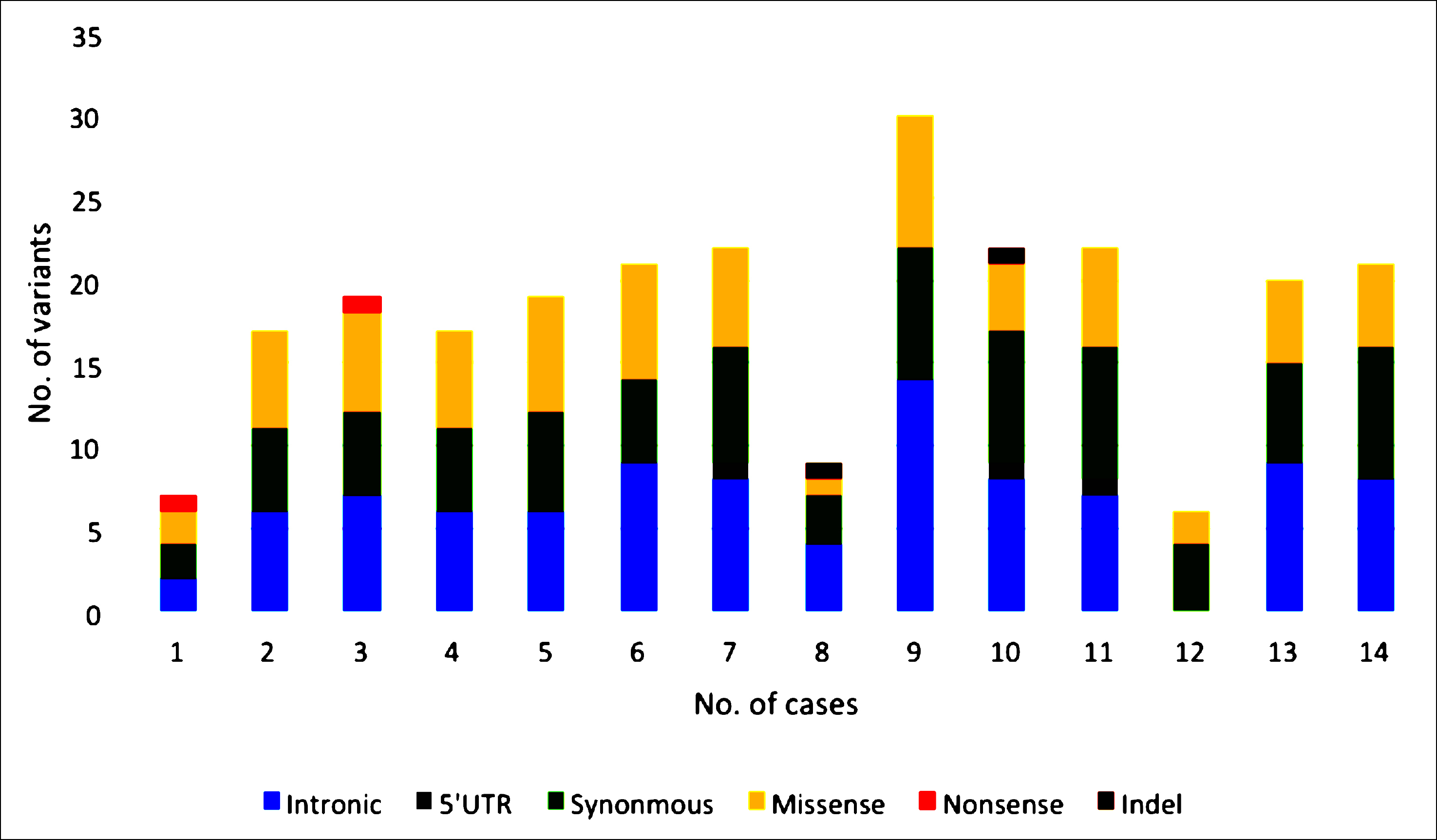 Figure 1 (A): Type of variants resulted from targeted NGS of BRCA panel in triple negative breast cancer and early onset breast cancer cases. There is an inframe indel in case # 8, and frameshift indel in case # 10.
Figure 1 (A): Type of variants resulted from targeted NGS of BRCA panel in triple negative breast cancer and early onset breast cancer cases. There is an inframe indel in case # 8, and frameshift indel in case # 10.
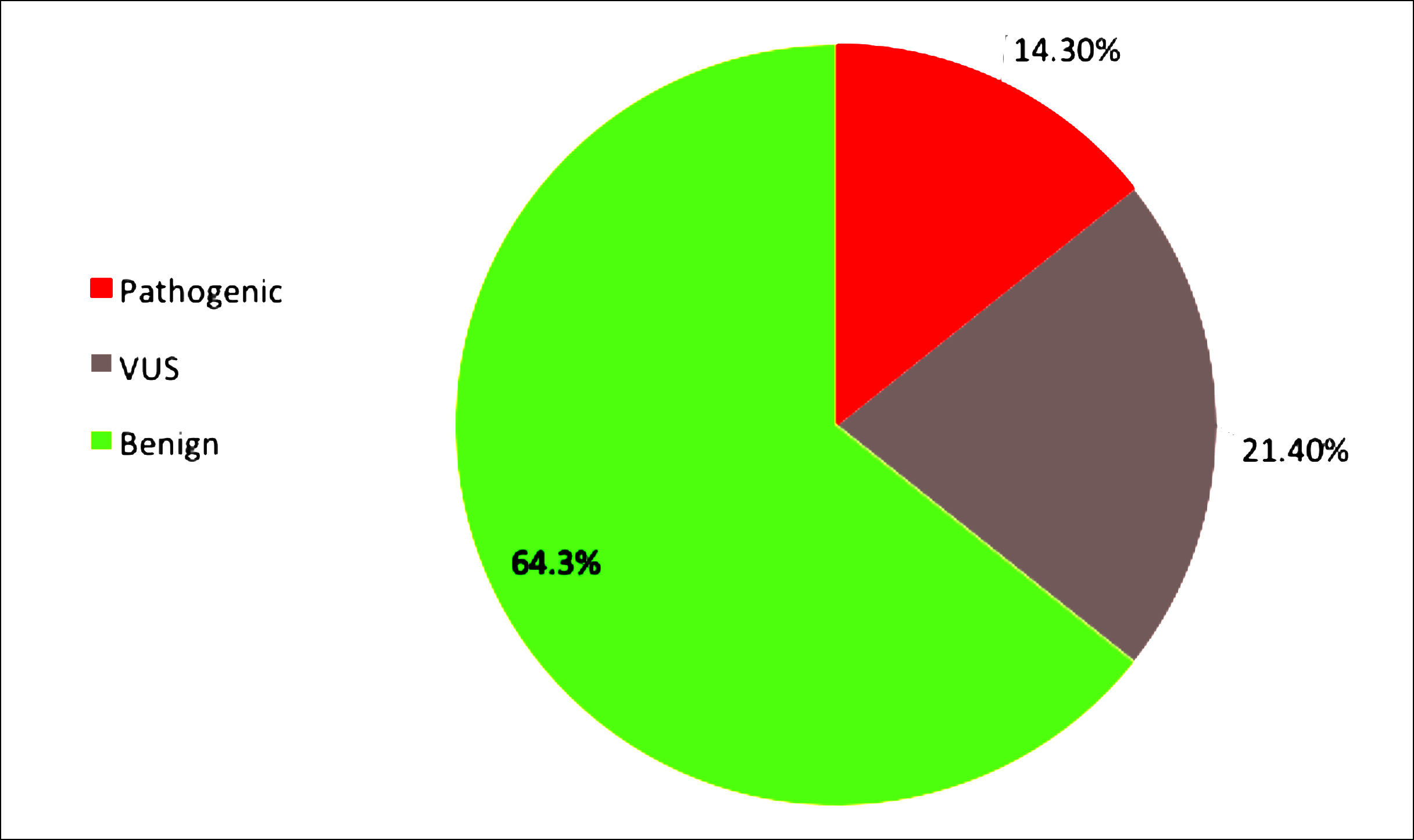 Figure 1 (B): Clinical significance of resulted BRCA variants in the cases under study. Two women with oncogenic variants (BRCA1), three with variant of uncertain significance (BRCA1 and BRCA2), and nine women with benign variants of BRCA genes reported in this study. Thus, providing a great deal of genetic information to their families.
Figure 1 (B): Clinical significance of resulted BRCA variants in the cases under study. Two women with oncogenic variants (BRCA1), three with variant of uncertain significance (BRCA1 and BRCA2), and nine women with benign variants of BRCA genes reported in this study. Thus, providing a great deal of genetic information to their families.
RESULTS
All fourteen female study participants had onset of disease at or less than 60 years age, and disease phenotype was invasive breast carcinoma with triple negative progesterone, estrogen and HER2 receptors.
In all patients’ BRCA1 and BRCA2 DNA sequences, 95% of bases had a phred score higher than 30, with the minimum 500X coverage depth, and the uniformity of coverage was 99%. There were 254 variants having variant quality score of 100, 64.8% and 48.4% variants in BRCA1 and BRCA2, respectively, in all patients under study. Consequence of variants were missense, nonsense, indels, and synonymous in exonic region and also detected variants in 5’UTR and splice site regions. Variants having minor allele frequency >0.05 based on gnomAD browser were excluded (Figure 1).
Table I: Details of pathogenic variants and VUS in BRCA1/BRCA2 resulted in cases under study by targeted NGS.|
Case’s Age |
Gene |
rsIDa |
Variantb |
MAFc |
Zygosity |
Consequence |
in silico Predictionf |
Classificationg |
|
31 |
BRCA1 |
rs41293455 |
Exon: 12/23 c.4327C >T p.(Arg1443Ter) |
Global= 0.000025, SAd= 0.000 |
Heterozygous
|
Stop gained/ Nonsense |
Disease causing |
Pathogenic |
|
38 |
BRCA1 |
rs775417240 |
Exon: 23/23 c.5571_5579delGATCCCCCA p.(Gln1857_Pro1859del) |
- |
Heterozygous
|
Inframe deletion |
Tolerated |
VUS |
|
32 |
BRCA1 |
rs80357819 |
Exon: 10/23 c.2866_2870delTCTCA p.(Ser956ValfsTer13) |
- |
Heterozygous
|
Frameshift deletion |
Disease causing |
Pathogenic |
|
45 |
BRCA2 |
rs587781399 |
Exon: 10/27 c.1070A>C p.(Glu357Ala) |
Global= 0.00004414, SA= 0.000365 |
Heterozygous
|
Missense |
Disease causing |
VUS |
|
33 |
BRCA2 |
rs431825354 |
Exon: 16/27 c.7628A>G p.(Tyr2543Cys) |
Global= 0.00003189, SA=0.000196 |
Heterozygous |
Missense |
Tolerated |
VUS |
|
rsID is reference SNP (RefSNP) identifier for a variant type assigned by Single Nucleotide Polymorphism Database (dbSNP), (https://www.ncbi.nlm.nih.gov/snp/ ). b Variant nomenclature according to HGVS (Human Genome Variation Society) standards. c Minor allele frequency (MAF) indicates the least frequent allele at a specific locus in a given population. For MAF, genome aggregation database (https://gnomad.broadinstitute.org/) was use. d SA is abbreviated form of South Asian. f Analysed by using in-silico prediction software i.e. Mutation Taster, SIFT and PolyPhen. g Interpretation was done by utilizing ACMG guidelines, curated external databases and resources i.e. dbSNP, ClinVar, GDC Data portal. |
||||||||
Two pathogenic variants (0.79%) were found in BRCA1, in cases aged less than 33 years. Variants of uncertain significance (1.18%) in BRCA1 and BRCA2 in three individual patients aged less than 46 years (Table I); and 98% variants in BRCA genes were benign in all cases.
DISCUSSION
Cancer genetic information is valuable for the patient’s targeted therapy and the family members to early diagnose and reduce the potential risk of cancer onset through proper genetic counselling and genetic testing. This study was conducted to establish a high-throughput targeted DNA sequencing focusing on BRCA1/2 genetic variants in triple-negative breast cancer cases. In a patient aged 31, a nonsense point mutation is detected which is causing a premature, shortened and non-functional BRCA1 protein. This variant has been reported to be causative of hereditary breast and ovarian cancer (dbSNP ID rs41293455), and was first reported in 1994 in Nature Genetics for the early onset of hereditary breast carcinoma.19 In a largest Pakistani study, it is found to be a germline deleterious variant for breast/ovarian cancer families.15 Another curated oncogenic BRCA1 variant, a frameshift deletion, has been detected in a case aged 32 (dbSNP ID rs80357819). This variant is creating a premature translational stop signal and has been reported in the literature for breast and/or ovarian cancer.20, 21
A variant of uncertain clinical significance (VUS) of BRCA1 gene has been detected (dbSNP ID rs775417240) in a case aged 38, an inframe deletion in the last exon of BRCA1. Interpretation of this variant in external databases is evaluated as of uncertain significance. There is no published paper which reports its pathogenicity for breast cancer and the submitters of this variant in ClinVar (ID: VCV000187111.6) are clinical laboratories. Prediction algorithm of Mutation Taster defines this variant as polymorphism which can be tolerated but experimental studies are not yet available to validate its clinical significance. Two VUS in BRCA2 gene have also been detected in the cases aged 45 and 33 respectively. Both of these variants are single base substitutions which are subsequently changing the amino acid and the consequence is missense variation. Although, these variants are reported in external databases and in a Pakistani study too,22 but there is no evidence for these variants to be causative of breast cancer. Further functional studies will be required to evaluate the roles of these variants of uncertain significance in disease phenotype as available evidence is not enough.
In this study, cases aged less than 50 years screened out to have reportable BRCA variants, were interpreted according to the ACMG guidelines, complementing other ethnic cohort studies. In a German consortium study, it was concluded that every triple negative breast cancer patient aged less 50 years should be screened for germline BRCA mutations regardless of family history.23 This assay is also supporting the literature that BRCA1 variants are more frequently associated with TNBC than BRCA2 variants.14
Other study participants in which benign mutations are identified, BRCA1/BRCA2 coding regions are ruled out to be causative for the germline breast cancer. Variants in other oncogenes or tumor suppressor genes might be responsible for their phenotype of triple negative breast cancer or it could be possible that no germline mutation is present in them and somatic variants might account for the cancer predisposition.
CONCLUSION
Understanding the genetic heterogenicity of breast cancer will help the clinicians to manage and treat it. PARP inhibitors are on the forefront approved targeted therapy against BRCA mutated carcinomas. Identification of these mutations will not only help identifying the most prevalent mutations in this region, but will also help introduce latest targeted therapy in these patients resulting in not only improved prognosis and treatment for cancer patients, but will also reduce the unwanted complications of traditional chemotherapy.
FUNDING:
Grateful to Armed Forces Institute of Pathology, RWP, Pakistan for providing us resources and laboratory space to conduct the experimentation, included in this study.
PATIENTS’ CONSENT:
Patients were interviewed and consents for genetic testing had been taken before the execution of this study.
CONFLICT OF INTEREST:
The authors declared no conflict of interest.
AUTHORS’ CONTRIBUTION:
HT, AG: Jointly conceived the study, designed and executed NGS panel in wet lab.
MZ: Integrated the NGS data and its compilation.
SRJ: Supervised and edited the manuscript.
NZ, GS: Edited the bio stats, took part in literature review and manuscript editing.
REFERENCES
- GLOBOCAN. Fact sheets by Cancer-GLOBOCAN 2018. https://gco.iarc.fr/today/data/factsheets/populations/586-pakistan-fact-sheets.pdf (Accessed on 14/11/2019).
- Boyle P. Triple-negative breast cancer: Epidemiological considerations and recommendations. Ann Oncol 2012; 23(suppl_6):vi7-12. doi: 10.1093/annonc/mds187.
- Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 2014; 343(6178):1470-5. doi: 10.1126/science.1252230.
- Chen H, Wu J, Zhang Z, Tang Y, Li X, Liu S, et al. Association between BRCA status and triple-negative breast cancer: A meta-analysis. Front Pharmacol 2018; 9:909. doi: 10.3389/fphar.2018.00909.
- NCCN Flash Updates: NCCN Guidelines® Updated www.nccn.org/about/news/ebulletin/ebulletindetail.aspx?ebulletinid=535 (Accessed on 2/10/2019).
- Vidula N, Bardia A. Targeted therapy for metastatic triple negative breast cancer: The next frontier in precision oncology. Oncotarget 2017; 8(63):106167. doi: 10.18632/ oncotarget.22580.
- Taylor KN, Eskander RN. PARP inhibitors in epithelial ovarian cancer. Recent Pat Anticancer Drug Disc 2018; 13(2):145-58. doi: 10.2174/1574892813666171204094822.
- Beniey M, Haque T, Hassan S. Translating the role of PARP inhibitors in triple-negative breast cancer. Oncoscience 2019; 6(1-2):287. doi: 10.18632/oncoscience.474.
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266(5182):66-71. doi: 10.1126/science.7545954.
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995; 378(6559):789-92. doi: 10.1038/ 378789a0.
- Roa BB, Boyd AA, Volcik K, Richards CS. Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nature genetics 1996; 14(2):185-7. doi: 10.1038/ ng1096-185.
- Janavičius R. Founder BRCA1/2 mutations in the Europe: Implications for hereditary breast-ovarian cancer prevention and control. EPMA J 2010; 1(3):397-412. doi: 10.1007/ s13167-010-0037-y.
- Copson ER, Maishman TC, Tapper WJ, Cutress RI, Greville-Heygate S, Altman DG, et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): A prospective cohort study. The Lancet Oncol 2018; 19(2):169-80. doi: 10.1016/S1470-2045(17)30891-4.
- Tun NM, Villani G, Ong K, Yoe L, Bo ZM. Risk of having BRCA1 mutation in high‐risk women with triple‐negative breast cancer: A meta‐analysis. Clin Genet 2014; 85(1): 43-8. doi: 10.1111/cge.12270.
- Rashid MU, Muhammad N, Bajwa S, Faisal S, Tahseen M, Bermejo JL, et al. High prevalence and predominance of BRCA1 germline mutations in Pakistani triple-negative breast cancer patients. BMC cancer 2016; 16(1):673. doi: 10.1186/s12885-016-2698-y.
- Wolff AC, Hammond ME, Allison KH, Harvey BE, Mangu PB. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. J Clin Oncol 2018; 142(11):1364-82. doi: 10.1200/JCO.2018.77.8738.
- Allison KH, Hammond ME, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL, et al. Estrogen and progesterone receptor testing in breast cancer: American society of clinical oncology/college of American pathologists guideline update. Archives Pathol Lab Med 2020; 144(5):545-63. doi: 10.5858/arpa.2019-0904-SA.
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015; 17(5):405-23. doi: 10.1038/gim.2015.30.
- Castilla LH, Couch FJ, Erdos MR, Hoskins KF, Calzone K. Mutations in the BRCA1 gene in families with early-onset breast and ovarian cancer. Nature genetics 1994; 8(4):387-91. doi: 10.1038/ng1294-387.
- Sun J, Meng H, Yao L, Lv M, Bai J, Zhang J, et al. Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients. Clinical Cancer Res 2017; 23(20):6113-9. doi.org/10.1158/1078-0432.CCR- 16-3227.
- Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Human Mutation 2018; 39(5): 593-620. doi.org/10.1002/humu.23406.
- Ahmad J, Le Calvez-Kelm F, Daud S, Voegele C, Vallee M, Ahmad A, et al. Detection of BRCA1/2 mutations in breast cancer patients from Thailand and Pakistan. Clinical Genet 2012; 82(6):594-8. doi: 10.1111/j.1399-0004.2012.01869.x.
- Engel C, Rhiem K, Hahnen E, Loibl S, Weber KE. Prevalence of pathogenic BRCA1/2 germline mutations among 802 women with unilateral triple-negative breast cancer without family cancer history. BMC Cancer 2018; 18(1):265. doi: 10.1186/s12885-018-4029-y.