Introduction of Human Leukocyte Antigen Typing by Next Generation Sequencing in Pakistan
By Hamid Nawaz Tipu, Muhammad NomanAffiliations
doi: 10.29271/jcpsp.2020.09.994Sir,
Introduction of next generation sequencing (NGS) technologies into clinical practice has enabled clinicians to utilise its potential in identifying genetic variations in various human disorders; and thus use targeted therapy based on personalised genome responses.1 World’s leading corporations in NGS technologies like Illumina, Thermo Fisher Scientific and BGI Genomics have introduced panels for various type of cancers, immune response and transplant genes. Human leukocyte antigen (HLA) gene typing carries critical significance in bone marrow and renal transplants, with outcome heavily dependent upon level of matching between donor and recipient.2 Typing HLA by NGS carries certain advantages over conventional sanger sequencing and sequence specific primers (SSP) based amplification. It resolves phasing problems and types up to eight digits’ resolution by sequencing millions of reads in parallel;3 thus, improving level of matching required in haploidentical or out of family transplants.
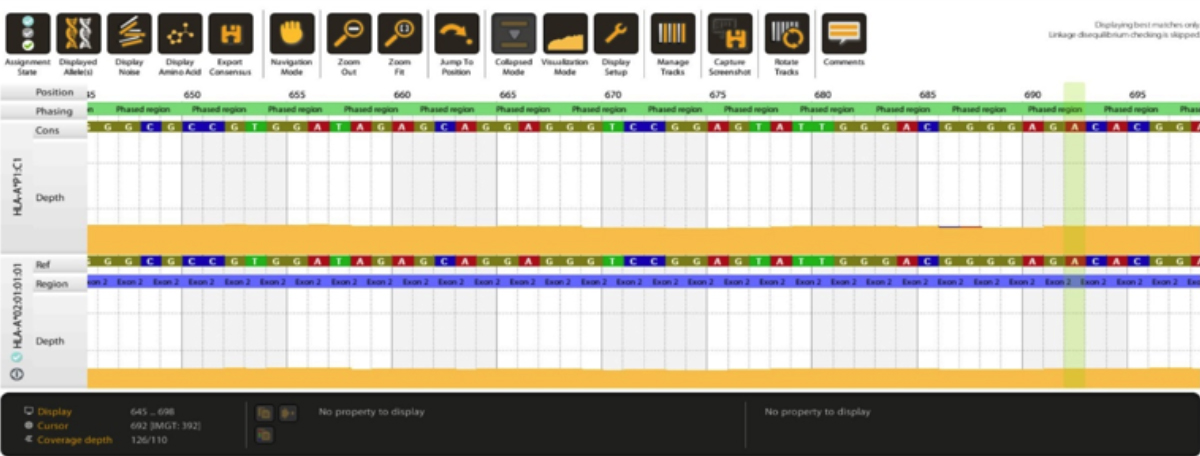 Figure1: Computing of a sample to HLA-A*02:01:01:01. Top DNA sequence shows reference sequence and bottom DNA sequence shows allele sequence.
Figure1: Computing of a sample to HLA-A*02:01:01:01. Top DNA sequence shows reference sequence and bottom DNA sequence shows allele sequence.
We have recently carried HLA typing by NGS, which was the first HLA typing by this technique in Pakistan to the best of our knowledge. Eighteen samples with known two-digit HLA SSP typing (Onelambda, USA) for HLA-A, B, C, DRB1 and DQB1 loci were included. For NGS, additionally, HLA-DPB1 and DQA1 loci were also typed. Locus specific amplification, library preparation, and size selection were all done using HLA holotype v3.0 (Omixon, Hungary), according to manufacturer’s instructions. Bioinformatic analysis for allele computing was done with HLA twin software version 3.1.1 (Omixon, Hungary), using consensus and statistical genotyping algorithms and database version 3.37.0_8. We were able to type all the 18 samples to their 7 loci (126 loci total) up to either 8-digit or up to 6-digit level (NMDP G group, wherever allele typing resolved to outside amplification range). Figure 1 shows results from allele 1 computing of a subject to 8-digit level. All the NGS quality parameters like Q30 and sample quality parameters like read count and coverage depth were above the minimum recommended levels.
Introduction of high resolution 6/8-digit typing is expected to significantly improve HLA matching accuracy between patients and their prospective donors.4 Its maximum utility lies in less than full match bone marrow transplants and establishment of donor’s registry to enhance donor pool in long term.
CONFLICT OF INTEREST:
Authors declared no conflict of interest.
AUTHORS’ CONTRIBUTION:
HNT: Conception and design of work, acquisition, analysis and interpretation, drafting of content and final approval, agreed to be accountable.
MN: Design of work, acquisition and analysis of work, agreed to be accountable.
REFERENCES
- Tipu HN, Shabbir A. Review article: evolution of dna sequencing. JCPSP 2015; 25(3):210-5.
- Allen ES, Yang B, Garret J, Ball ED, Maiers M, Morris GP. Improved accuracy of clinical HLA genotyping by next-generation DNA sequencing affects unrelated donor search results for hematopoietic stem cell transplantation. Hum Imm 2018; 79(12):848-54.
- Duke JL, Lind C, Mackiewicz K, Ferriola D, Papazoglou A, Gasiewski A et al. Determining performance characteristics of an NGS-based HLA typing method for clinical applications. HLA 2016; 87(3):141-52.
- Smith AG, Pereira S, Jaramillo A, Stoll ST, Khan FM, Berka N et al. Comparison of sequence‐specific oligonucleotide probe vs next generation sequencing for HLA‐A, B, C, DRB1, DRB3/B4/B5, DQA1, DQB1, DPA1, and DPB1 typing: Toward single‐pass high‐resolution HLA typing in support of solid organ and hematopoietic cell transplant programs. HLA 2019; 94(3):296-306.