Comparison of Sequence Specific Primers in the Next Generation Sequencing in Human Leukocyte Antigen Typing for Transplant Recipients
By Hamid Nawaz TipuAffiliations
doi: 10.29271/jcpsp.2020.11.1138ABSTRACT
Objective: To compare two human leukocyte antigen (HLA) typing methods, namely sequence specific primers (SSP) and next generation sequencing (NGS) for alleles concordance
Study Design: Descriptive study.
Place and Duration of Study: Immunology department, Armed Forces Institute of Pathology, Rawalpindi, Pakistan, from December 2019 to May 2020.
Methodology: A total of 48 subjects, including 30 males and 18 females, were typed by NGS at 7 loci, making a total of 672 types loci. SSP typing was done for 276 loci among these. Comparison was made at SSP level of low resolution. NGS typing was done with Illumina’s MiSeq instrument using Omixon HLA holotype 7 loci kit and analysis done with HLA twin software. SSP typing was done with micro SSP kit from onelambda. Statistical analysis was done using statistical package for social sciences (SPSS) version 24.0.
Results: Among the 672 NGS types loci and 276 SSP types loci, there were mismatches at one B locus and one C locus, whereby NGS computed HLA-B*58:01 and HLA-C*12:02 while SSP detected HLA-B*57 and HLA-C*05, respectively. At remaining 274 loci, HLA typing fully matched at low resolution, making concordance rate 99.3%. Commonest alleles detected by NGS were HLA-A*02:01, B*51:01, C*07:02, DPB1*04:01, DQA1*01:03, DQB1*02:01 and DRB1*13:01.
Conclusion: Batch testing, high throughput, improved accuracy, more loci coverage, maximum gene coverage including all exons and introns and high-resolution typing confer significant advantages to next generation sequencing over old methods of HLA typing. This technique is suitable for high throughput laboratories. High running cost hampers its routine implementation in 3rd world countries.
Key Words: Human leukocyte antigen, Next generation sequencing, High resolution.
INTRODUCTION
Human Leukocyte Antigens (HLA) are the most polymorphic loci in human genome. Due to extremely high immunogenicity, HLA matching among transplant recipients and donors is associated with significantly better outcome.1 Moreover, these find utility in human population genetic studies,2 disease association3 and pharmacogenomic.4 British Society for Histocompatibility and Immunogenetics (BSHI) guidelines suggest that patient and donor for haematopoietic progenitor cell transplantation should be matched for at least HLA A, B, C, DRB1 and DQB1 loci by a deoxyribose nucleic acid (DNA)-based method including high resolution typing.5
The extent of HLA match significantly impacts transplant outcome, although it may depend upon several other factors like patient’s age, primary disease, graft source and conditioning regimen.6
Advancements in technology have led to most of the laboratories shifting from serological- based methods to molecular methods. Currently, sequence specific primers (SSP) and sequence specific oligonucleotide probes (SSOP) are the routinely used DNA-based methods. These techniques provide rapid turnaround time, but can resolve HLA up to 2 digits typing only, making them unsuitable for less than full match transplant and across family transplants. Sanger sequencing is the gold standard providing high resolution up to 4 digits typing, but it only interrogates polymorphic exons 2 and 3 for HLA class I and exon 2 for HLA class II. It also does not resolve phase ambiguities which may culminate into ambiguous typing.7,8 Next generation sequencing (NGS) technologies have evolved tremendously over last decade and as such, have found their way in HLA typing also. Despite high initial cost, NGS technologies offer high throughput (by batch testing), more HLA loci typing, high typing confidence (coverage depth), highest possible resolution (up to 8 digits) and reduced cost (when multiplexed).9 HLA typing by NGS methods has clearly been shown to improve unambiguous genotyping when compared to conventional methods.10 Moreover, stem cell transplant recipients, who are HLA typed by ultra-high resolution NGS methods, have been shown to have superior survival.11
HLA NGS typing has yet little penetration in the Third World countries. To authors knowledge, their centre is the first one to offer this technique to transplant recipients in Pakistan. With a population of over 220 million and inhabitants from different ethnic groups, HLA polymorphisms are expected to find extreme genetic diversity in Pakistan. This study was undertaken to compare HLA typing by NGS to the existing technique (sequence specific polymorphism, SSP), in terms of throughput, turnaround time, cost analysis, accuracy, loci typed and typing confidence; and to comprehensively analyse the quality parameters applied in HLA NGS typing to ensure that high quality typing is available to patients with minimum chances of error.
METHODOLOGY
This descriptive study was carried out in Immunology Department of Armed Forces Institute of Pathology, Rawalpindi, Pakistan, from December 2019 to May 2020, after approval from Institution’s Ethical Committee. A total of 48 subjects were included in the study by non-probability convenience sampling after their informed consent. All the patients of haematopoietic stem cell transplant (HSCT) coming for HLA typing, during the study period, were included along with their extended family of siblings and parents (as donors) wherever available. All these subjects were HLA typed by NGS at 7 loci, making a total of 672 typed HLA loci (each locus co-dominant). Forty-two of these subjects (28 males and 14 females) were also initially typed by SSP method at 276 loci.
HLA NGS typing was carried out on Illumina MiSeq instrument (Illumina, USA) using Holotype HLA 24/7 version 3.0 kit (Omixon, Hungry), according to manufacturer’s recommendations. This is a new and extensive procedure and is briefly mentioned here. Genomic DNA was extracted (Qiagen, USA) and concentration adjusted to 25 ng/ul fluorometrically (Qubit, USA) using Qubit dsDNA HS assay kit (Thermo fisher, USA). HLA was amplified at 7 loci as recommended by Holotype kit manufacturer. Since amplicon size ranged 3-7000 kilo bases (kb), amplification was verified on 2% agarose gel. This was followed by amplicon quantitation (Qubit, USA), dilution to adjust final concentration to 50-100 ng/ul, amplicon pooling and purification with ExoSAP-IT (Thermo fisher, USA). Library was prepared by enzymatically fragmenting amplicons, end repairing and ligating with indices and adapters. Size selection of amplicons was done with Agencout AMPure XP beads (Beckman Coulter, USA). After final library quantification and concentration adjustment, sequencing was started on MiSeq instrument (Illumina, USA) using micro 300 flow cell. Protocol was set to Fastq file generation. Bioinformatic analysis was done on Omixon HLA Twin v 3.1.3 software that is dual algorithm genotyping software and uses both consensus genotyping and statistical genotyping independently. Final HLA genotyping assignment was done at 4-digit level (high resolution) after taking into account HLA type of whole family and haplotype construction.
HLA SSP typing was carried out using One Lambda micro SSP kit for A, B, C, DRB1 and DQB1 (One Lambda, USA) and results were analysed using HLA Fusion software version 4.3. Data for HLA typing was entered in Statistical Package for Social Sciences (SPSS) version 24 (IBM Corp, Armonk, NY) and analysed for frequencies, percentage, mean, SD and concordance.
Quality control of NGS typing was done with Omixon HLA Twin version 3.1.3 software for every sample and locus individually. Primary quality control (QC) parameters were analysed including read count, noise ratio, key exon spot noise ratio, consensus coverage key exon minimum depth, key exon allele imbalance and genotype available. Secondary QC parameters included fragment size, read quality, other exon spot noise ratio, PCR crossover artifact ratio and key exon mismatch count. Omixon HLA twin traffic light system, provided by software manufacturer, was applied to view results marked for compromised QC metrices.
RESULTS
There were total 30 males and 18 females. Over 6 months, 7 loci (HLA-A, B, C, DPB1, DQA1, DQB1, DRB1) of 48 subjects were typed by NGS. Due to Mendelian inheritance and co-dominant expression, each locus has two alleles in each individual, this makes a total of 672 types loci in 48 individuals. It included 30 males (62.5%) and 18 females (37.5%) with a mean age 27.7+16.7 years. Comparison was made at 276 loci typed with SSP, and at 2-digit level (low resolution). There were mismatches at one B locus and one C locus, whereby NGS computed HLA-B*58:01 and HLA-C*12:02, while SSP detected HLA-B*57 and HLA-C*05, respectively. At remaining 274 loci, HLA typing fully matched at low resolution achieved by SSP, making concordance rate 99.3%. Table I shows concordance rate among individual loci for both NGS and SSP methods. Table II shows top three most common HLA alleles at high resolution 4-digit level, detected by NGS method. Table III shows comparison of two methods in terms of cost, turnaround time, throughput, loci tested and resolution. Please note that these parameters are those as determined in our laboratory, each laboratory might have variations from it.
Figure 1 shows the analysis of primary and secondary QC parameter indices for an analysed NGS sample. Green circle with numbers shows fully passed parameters.
All the QC parameters of this study fell in the range of passed, info, inspect and investigate, that successfully assigned genotyping by the HLA twin software. Only one primary QC parameter, consensus coverage key exon minimum depth, failed the criterion of minimum 10x coverage depth in two DRB1 alleles in two different samples in the same batch. However, the assigned genotype still matched the SSP alleles. Figure 2 shows the low coverage depth for assigned allele DRB1*11:84 at exon 2.
Table I: Loci typed by NGS and SSP and concordance among them.
|
Method |
HLA-A |
HLA-B |
HLA-C |
HLA-DPB1 |
HLA-DQA1 |
HLA-DQB1 |
HLA-DRB1 |
Total |
|
NGS |
96 |
96 |
96 |
96 |
96 |
96 |
96 |
672 |
|
SSP |
72 |
72 |
40 |
0 |
0 |
46 |
46 |
276 |
|
Concordance (%) |
72/72 (100) |
71/72 (98.6) |
39/40 (97.5) |
0/0 (0) |
0/0 (0) |
46/46 (100) |
46/46 (100) |
274/276 (99.3) |
Table II: Three commonest high-resolution alleles at each locus, as detected by NGS.
|
HLA locus |
Commonest alleles |
Frequency (n=96 per locus) |
Percentage (%) |
|
A |
02:01 |
19 |
19.8 |
|
11:01 |
18 |
18.8 |
|
|
01:01 |
9 |
9.4 |
|
|
B |
51:01 |
18 |
18.8 |
|
52:01 |
18 |
18.8 |
|
|
08:01 |
9 |
9.4 |
|
|
C |
07:02 |
18 |
18.8 |
|
12:02 |
14 |
14.6 |
|
|
15:02 |
11 |
11.5 |
|
|
DPB1 |
04:01 |
35 |
36.5 |
|
02:01 |
13 |
13.5 |
|
|
126:01 |
9 |
9.4 |
|
|
DQA1 |
01:03 |
28 |
29.2 |
|
05:01 |
19 |
19.8 |
|
|
01:02 |
13 |
13.5 |
|
|
DQB1 |
02:01 |
19 |
19.8 |
|
06:03 |
18 |
18.8 |
|
|
05:03 |
10 |
10.4 |
|
|
06:01 |
10 |
10.4 |
|
|
DRB1 |
13:01 |
18 |
18.8 |
|
03:01 |
15 |
15.6 |
|
|
14:04 |
10 |
10.4 |
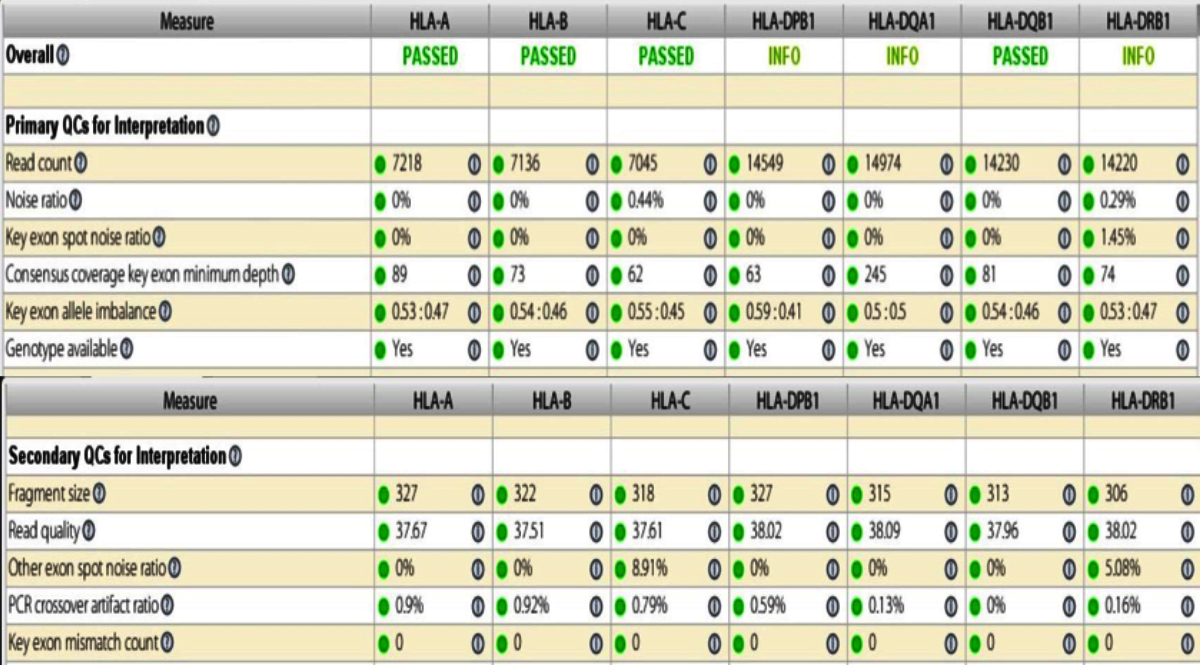 Figure 1: Analysis of primary and secondary QC parameters for an NGS sample. All values with green circles indicate passed parameter.
Figure 1: Analysis of primary and secondary QC parameters for an NGS sample. All values with green circles indicate passed parameter.
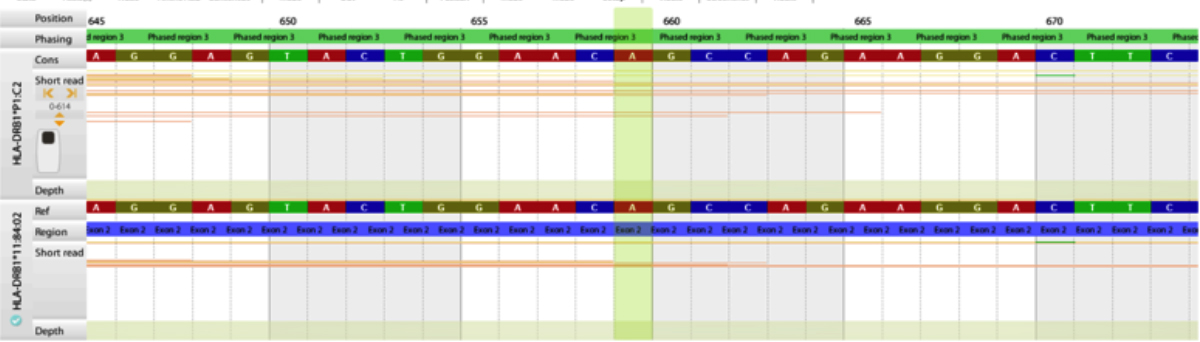 Figure 2: Low coverage depth (X) for DRB1*11:84 at exon 2.
Figure 2: Low coverage depth (X) for DRB1*11:84 at exon 2.
DISCUSSION
HLA matching between transplant recipients and their donors has long been associated with improved outcomes. A single HLA mismatch has found to be associated with only 13% increased risk of kidney graft failure; whereas, six HLA mismatch cause 64% increase in risk of graft failure.12 Similarly, in bone marrow transplants, main criterion of donor selection is HLA compatibility between donor and recipient.13 Molecular methods have greatly improved our understanding and thus extent of matching in transplants.14 Several commercial vendors have introduced their HLA NGS typing instruments and techniques which have drastically improved outcomes of HLA typing in different areas like transplantation, population genetics and disease association studies.15
In the current study, we evaluated HLA NGS typing Omixon Holotype HLA 24/7 kit for introduction of high-resolution typing facility to these transplant recipients and donors. We measured its concordance at low resolution with previously established SSP method. For NGS, genotype assignment was considered valid only when all the QC parameters met minimum passing criterion by the manufacturer. For two DRB1 alleles of two different samples in same batch, key exon minimum coverage depth, that is commonly referred to as coverage depth, was below minimum criterion of 10x. However, since it matched SSP assigned allele, thus genotype assignment was considered valid and included in concordance analysis. The concordance between two methods was 99.3%, with only one HLA-B and one HLA-C alleles differing. HLA-B*58:01 computed with NGS has 99% sequence similarity (1073/1089 identities) to HLA-B*57:01, as determined by National Centre of Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST). It is possible that due to lower read count, NGS method might have mistaken the correct allele. HLA-C*12:02 determined by NGS has 99% (1088/1101 identities) sequence similarity to C*05:01, however, all the quality parameters for NGS were correct. So quite possibly, SSP method might have determined the wrong allele here. Gowda et al. compared Roche 454 NGS sequencing technology to SSP/SSOP and have found over 95% concordance between two methods, though their sample size was slightly larger than the presently reported.16 Smith et al. have measured 99% concordance between NGS and SSOP, on 289 samples.17
HLA is the most polymorphic region in human genome and different populations carry different HLA genetic background; which in turn, may affect their disease association and infectious susceptibility.18
Table III: Comparison of two methods for laboratory functioning parameters.|
|
Throughput per batch (samples) |
Turnaround time (days) |
Hand on time (hours) |
Cost per test (USD approx.) |
Loci tested (minimum) |
Resolution (digits) |
|
NGS |
12-24 |
3 |
3-4 |
250 |
7 |
4-8 |
|
SSP |
1-2 |
1 |
2-3 |
100 |
5 |
2 |
Although the sample size was small for population studies but commonest alleles were HLA-A*02:01, B*51:01, C*07:02, DPB1*04:01, DQA1*01:03, DQB1*02:01 and DRB1*13:01. Gowda et al. have found HLA-A*24:02, B*40:06, C*04:01, DQB1*03:01 and DRB1*07:01 to be commonest alleles on cohort of 80 samples.16
Finally, the authors compared the two techniques for routine laboratory functioning parameters. One could run large number of samples in an NGS batch depending upon several factors. The authors ran 12-24 samples in different batches using Illumina’s MiSeq instrument and micro flow cell. In SSP, although one could process several samples together but it significantly increases workload complexity and increases chances of error. Therefore routinely, not more than 2 samples are processed for SSP at a time. However, SSP is quick to provide results on the same day, while combining all the work flow of NGS together, results are usually available on the third day. Hence, for a busy laboratory with high throughput, NGS is a suitable option. While for a smaller laboratory when only few samples are required to be typed or in urgent situations, SSP may be suitable alternative, although, it depends upon several other factors like level of resolution required and finances. The running cost of NGS is more than double of SSP (USD 250 vs. 100 approx.) although this difference can be reduced by using smaller nano flow cell. Cost also depends upon country’s laws, taxes and supplier’s profits. NGS typing definitely carries significant advantage of testing several loci covering whole of the gene instead of just the key exons. In our study we typed 7 loci but options for more loci are also available in market. High resolution typing available with NGS is rapidly becoming the gold standard for bone marrow as well as renal transplants19 and it is expected to outdate SSP in coming few years.
CONCLUSION
High resolution HLA typing by NGS offers a forward approach for transplant recipients for improving transplant outcomes. It is rapidly becoming the gold standard and has high concordance (over 99%) with previously established SSP. Batch testing, high throughput, improved accuracy, more loci coverage, maximum gene coverage including all exons and introns and high-resolution typing confer it significant advantages over old methods of HLA typing. This technique could be employed in a large HLA typing laboratory with high throughput because currently high cost hampers its use for limited number of samples.
ACKNOWLEDGMENTS:
We are thankful to our Head of Department, Dr Dawood Ahmed and our laboratory in charge, Dr Raza Jaffer, for providing institutional support for the study. We are also grateful to our Institutional Ethical Review Committee for granting us permission over ethical aspects of the projects, under letter No. Cons/IMM-2/READ-IRB/20/479.
ETHICAL APPROVAL:
Ethical approval was obtained from Institutional Review Board of Armed Forces Institute of Pathology prior to initiation of work, under Letter No. Cons/IMM-2/READ-IRB/479 dated 22nd Oct 2019.
PATIENTS’ CONSENT:
Informed consents have been obtained from subjects that their data may be used for research purposes.
CONFLICT OF INTEREST:
The author declared no conflict of interest.
AUTHOR’S CONTRIBUTION:
HNT: Conception and design, data acquisition and analysis, data interpretation, article drafting and revising, final approval.
REFERENCES
- Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007; 110(13):4576–83. doi: 10.1182/blood-2007-06-097386.
- Sanchez-Mazas A, Meyer D. The relevance of HLA sequencing in population genetics studies. J Immunol Res 2014; 2014:971818. doi.org/10.1155/2014/971818.
- Holoshitz J. The quest for better understanding of HLA-disease association: Scenes from a road less travelled by. Discov Med 2013; 16(87):93-101.
- Pavlos R, Mallal S, Phillips E. HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 2012; 13(11): 1285-1306. doi: 10.2217/pgs.12.108.
- Little AM, Green A, Harvey J, Hemmatpour S, Latham K, March SGE, et al. BSHI Guideline: HLA matching and donor selection for haematopoietic progenitor cell transplantation. Int J Immunogenet 2016; 43(5):263-86. doi: 10.1111/iji.12282.
- Furst D, Neuchel C, Tsamadou C, Schrezenmeier H, Mytilineos J. HLA Matching in Unrelated Stem Cell Transplantation up to Date. Transfus Med Hemother 2019; 46(5): 326-36. DOI: 10.1159/000502263.
- De Santis D, Dinauer D, Duke J, Erlich HA, Holcomb CL, Lind C, et al. 16th IHIW: review of HLA typing byNGS. Int J Immunogenet 2013; 40(1):72-6. doi:10.1111/iji.12024.
- Tipu HN, Shabbir A. Evolution of DNA Sequencing. JCPSP 2015; 25(3):210-5.
- Lind C, Ferriola D, Mackiewicz K, Heron S, Rogers M, Slavich L, et al. Next-generation sequencing: The solution for high-resolution, unambiguous human leukocyte antigen typing. Hum Immunol 2010; 71(10):1033-42. doi: 10.1016/ j.humimm.2010.06.016.
- Allena ES, Yanga B, Garrettb J, Ballb ED, Maiersc M, Morrisa GP. Improved accuracy of clinical HLA genotyping by next-generation DNA sequencing affects unrelated donor search results for haematopoietic stem cell transplantation. Hum Immunol 2018; 79(12): 848-54. doi: 10.1016/j.humimm. 2018.10.008.
- Mayor NP, Hayhurst JD, Turner TR, Szydlo RM, Shaw BE, Bultitude WP, et al. Recipients receiving better HLA-matched haematopoietic cell transplantation grafts, uncovered by a novel hla typing method have superior survival: A retrospective study. Biol Blood Marrow Transplant 2019; 25(3):443-50. doi.org/10.1016/ j.bbmt.2018.12.768.
- Williams RC, Opelz G, McGravey CJ, Weil EJ, Chakkera HA. The risk of transplant failure with hla mismatch in first adult kidney allografts from deceased donors. Transplantation 2016; 100(5):1094-1102. doi: 10.1097/TP.000000 0000001115.
- Wang Y, Wu D-P, Liu Q-F, Xu L-P, Liu K-Y, Zhang X-H, et al. Donor and recipient age, gender and ABO incompatibility regardless of donor source: validated criteria for donor selection hematopoietic transplants. Leukemia 2018; 32: 492-8.
- Montgomery RA, Tatapudi VS, Leffell MS, Zachary AA. HLA in transplantation. Nat Rev Nephrol 2018; 14: 558-70.
- Carapito R, Radosavljevic M, Behram S. Next-generation sequencing of the hla locus: Methods and impacts on HLA typing, population genetics and disease association studies. Hum Immunol 2016; 77(11): 1016-23. doi: 10.1016/ j.humimm.2016.04.002.
- Gowda M, Ambardar S, Dighe N, Manjunath A, Shankaralingu C, Hirannaiah P, et al. Comparative analyses of low, medium and high-resolution HLA Typing technologies for human populations. J Clin Cell Immunol 2016; 7(2): 0.4172/2155-9899.1000399.
- Smith AG, Pereira S, Jaramillo A, Stoll ST, Khan FS, Berka N, et al. Comparison of sequence-specific oligonucleotide probe vs next generation sequencing for HLA-A, B, C, DRB1, DRB3/B4/B5, DQA1, DQB1, DPA1, and DPB1 typing: Toward single-pass high-resolution HLA typing in support of solid organ and haematopoietic cell transplant programs. HLA 2019; 94(3):296-306. Doi.10.1111/tan.13619.
- Alter I, Gragert L, Fingerson S, Maiers M, Louzoun Y. Allelic resolution NGS HLA typing of class I and class II loci and haplotypes in Cape Town, South Africa. PLOS Computational Biology 2017; doi.org/10.1371/journal. pcbi.1005693.
- Melista E, Rigo K, Pasztor A, Christiansen M, Bertinetto FE, Meintjes P, et al. Towards a new gold standard- NGS corrections to sanger SBT genotyping results. Human Immunol 2015; 76. doi.org/10.1016/j.humimm.2015. 07.205.