Can Next-generation Sequencing Replace Fecal Immunochemical Tests or CT in the Screening of Colorectal Cancer and Advanced Adenoma?
By Xiaoling Ye, Hong Chai, Chunyan Huang, Miao Liu, Tao DengAffiliations
doi: 10.29271/jcpsp.2020.09.940ABSTRACT
Objective: To explore the feasibility of next-generation sequencing (NGS) for the screening of colorectal cancer (CRC) and advanced adenoma (AA).
Study Design: Observational study.
Place and Duration of the Study: Renmin Hospital of Wuhan University, Wuhan, China, from June 2019 to February 2020.
Methodology: Patients who met inclusion and exclusion criteria were divided into three groups: CRC, AA, and control group. Then, the gene methylation status in the blood in the CRC, AA, and control group was analysed by NGS, and the CRC screening risk assessment model was used for comprehensive analysis. Afterwards, the methylated haplotype index (PHF Index) was calculated, and the screening results of the patients were classified as positive or negative, according to the score. The clinicopathological results were used as the gold standard, and the screening results for NGS were compared with the computed tomography (CT) and fecal immunochemical test (FIT) to text its feasibility.
Results: NGS has a certain detection ability for CRC, with a sensitivity of 57.1% (8/14). This was higher than that of FIT and CT, and the combined positive rate of these three methods could reach 92.3% (12/13). The sensitivity of detection of AA could reach up to 75.0% (6/8) after combining with FIT and CT. The positive rate of the NGS test for postoperative CRC was 23.1% (3/13), which was significantly lower than preoperative CRC. The sensitivity of CT for preoperative CRC detection was only 45.5% (5/11), but the specificity could reach up to 98.2% (55/56), which was higher than NGS (71/78, 91.0%) and FIT (27/33, 81.8%).
Conclusion: Although NGS cannot replace FIT and CT at present, this provides a new effective and auxiliary detection method for people who are unsuitable or unwilling to receive colonoscopy.
Key Words: Colorectal neoplasms, Adenoma, High-throughput nucleotide sequencing, Mass screening, Fecal immunochemical tests, Computed tomography.
INTRODUCTION
Colorectal cancer (CRC) is a common malignant gastrointestinal tumor, and its incidence is second among women and third among men. It also has a high mortality rate, and the age of onset tends to be younger.1-3 Colorectal advanced adenoma (AA) refers to adenoma with a diameter of more than 10 mm, and villous adenoma (villous component >25%) or high-grade intraepithelial neoplasia, which belongs to the category of precancerous lesions, and develops into invasive CRC, with an annual conversion rate of up to 2.6-5.7%.4
Colonoscopy is presently the gold standard for detecting CRC.5 However, high cost, invasive procedures, and the relatively high risk of complications make it difficult to widely achieve this, especially in some underdeveloped areas. In addition, the sensitivity and specificity of serum tumor markers are also low.6,7 Hence, biomarkers with high sensitivity, specificity, and compliance for CRC screening in blood are urgently needed for the early detection of CRC.
The main molecular mechanisms that lead to CRC include chromosomal instability (CIN), microsatellite instability (MSI), and CpG island methylation phenotype (CIMP).8 DNA methylation is an important epigenetic modification, and the CpG island is the main site of DNA methylation, which is closely correlated to the occurrence and development of tumors.9,10 The high methylation of the CpG island in the promoter region of the tumor suppressor gene is an important indicator of gene inactivation,11 and the increased promoter methylation frequency of the tumor suppressor gene exists in many tumor tissues. Hence, this may be used as one of the main indicators in the clinical diagnosis of CRC.
Table I: Comparative analysis of the positive rate of next-generation sequencing in each group (Independent t-test and Mann-Whitney test).|
Items |
Total number |
The average score (95%CI) |
Positive patients [n, (%)] |
comparative analysis of positive rates in each group |
p-value |
|
Preoperative CRC group |
14 |
10.807 (8.409-13.205) |
8 (57.1) |
Preoperative CRC group vs Normal group |
0.002 |
|
Postoperative CRC group |
13 |
9.285 (6.218-12.352) |
3 (23.1) |
Postoperative CRC group vs Normal group |
0.602 |
|
AA group |
15 |
7.640 (6.563-8.717) |
2 (13.3) |
AA group vs Normal group |
0.492 |
|
Common adenoma group |
27 |
7.511 (6.943-8.079) |
2 (7.4) |
Common adenoma group vs Normal group |
>0.999 |
|
Polyps group |
28 |
7.704 (6.871-8.536) |
4 (14.3) |
Polyps group vs Normal group |
0.309 |
|
Inflammation group |
12 |
7.825 (5.769-9.881) |
1 (8.3) |
Inflammation group vs Normal group |
>0.999 |
|
Normal group |
11 |
7.309 (6.429-8.189) |
0 (0.0) |
Preoperative CRC group vs AA group |
0.011 |
|
Total |
120 |
8.162 (7.627-8.697) |
20 (16.7) |
Preoperative CRC group vs postoperative CRC group |
0.109 |
|
Pre-CRC group: Preoperative CRC group; Post-CRC group: Postoperative CRC group; CRC: Colorectal cancer; AA: Advanced adenoma. The screening score greater than 10 is considered as positive. |
|||||
Table II: Comparative analysis of the positive detection rates of each group by NGS, FIT and CT.
|
Items |
Pre-CRC group (%) |
Post-CRC group (%) |
AA group (%) |
Control group (%) |
Pre-CRC group vs control group (p-value) |
Post-CRC group vs control group (p-value) |
AA group vs control group (p-value) |
Pre-CRC group vs AA group |
|
NGS |
8 (57.1) |
3 (23.1) |
2 (13.3) |
7 (9.0) |
<0.001 |
0.151 |
0.634 |
0.021 |
|
CT |
5 (45.5) |
4 (57.1) |
1 (7.7) |
1 (1.8) |
<0.001 |
<0.001 |
0.344 |
0.061 |
|
FIT |
7 (50.0) |
6 (46.2) |
5 (38.5) |
6 (18.2) |
0.037 |
0.070 |
0.248 |
0.547 |
|
NGS+CT |
11 (78.6) |
7 (77.8) |
3 (21.4) |
8 (13.8) |
<0.001 |
<0.001 |
0.438 |
0.002 |
|
NGS+FIT |
11 (78.6) |
7 (53.8) |
6 (46.2) |
12 (33.3) |
0.004 |
0.193 |
0.508 |
0.120 |
|
FIT+CT |
8 (88.9) |
7 (87.5) |
5 (71.4) |
7 (31.8) |
0.006 |
0.012 |
0.092 |
0.550 |
|
NGS+FIT+CT |
12 (92.3) |
8 (88.9) |
6 (75.0) |
13 (46.4) |
0.005 |
0.050 |
0.236 |
0.531 |
|
Pre-CRC group: Preoperative CRC group; Post-CRC group: Postoperative CRC group; CRC: Colorectal cancer; AA: Advanced adenoma; NGS: Next-generation sequencing; CT: Computerised tomography; FIT: Fecal immunochemical tests. |
||||||||
Table III: Summary of preoperative detection ability of preoperative CRC by different detection methods.
|
Items |
Sensitivity (%) |
Specificity (%) |
PPV (%) |
NPV (%) |
|
NGS |
57.1 |
91.0 |
53.3 |
92.2 |
|
CT |
45.5 |
98.2 |
83.3 |
90.2 |
|
FIT |
50.0 |
81.8 |
53.8 |
79.4 |
|
NGS+CT |
78.6 |
86.2 |
57.9 |
94.3 |
|
NGS+FIT |
78.6 |
66.7 |
47.8 |
88.9 |
|
FIT+CT |
88.9 |
68.2 |
53.3 |
93.8 |
|
NGS+FIT+CT |
92.3 |
53.6 |
48.0 |
93.8 |
|
PPV: Positive predictive value; NPV: Negative predictive value; CRC: Colorectal cancer; AA: Advanced adenoma; NGS: Next-generation sequencing; CT: Computerised tomography; FIT: Fecal immunochemical tests. |
||||
Recent studies have shown that the abnormal methylation of many genes has occurred in the early stages of CRC and precancerous lesions, and that this can be detected in the peripheral blood, feces, and other body fluids of patients.12 Single-gene-based DNA methylation biomarkers have limited sensitivity and specificity. Therefore, the simultaneous detection of methylation sites of multiple genes may be a good method.
The aim of the present study was to use next-generation sequencing (NGS) technology, which is a revolutionary change of traditional sequencing, has the advantages of high throughput, high sensitivity and high degree of automation to detect the multiple gene methylation of the CpG island in the blood of CRC patients, patients with AA and normal subjects, and explore the potential application value of NGS in CRC screening.
METHODOLOGY
The present study was performed in Renmin Hospital of Wuhan University from June 2019 to February 2020. Inclusion criteria were age >18 years; and patients who had underwent NGS, and signed an informed consent and volunteered to join the study. Exclusion criteria were pregnancy or lactation; severe trauma that had been treated with transfusion within the past week; patients who participated in "interventional" clinical trials, and received any experimental medicine within the past 30 days; patients who had neurological diseases, such as stroke and dementia or patients who had other cancers. These standards were developed to better exclude the effects of other substances (except the colorectal disease) in blood. The study was approved by the Ethics Committee of the Hospital. All procedures performed in the present study, which involved human participants, were conducted in accordance to the ethical standards of the institutional research committee, and with the 1964 Helsinki declaration and its later amendments. All patients provided a signed informed consent prior to testing.
Blood was drawn after one week of withdrawal, when the patient was taking anticoagulant drugs, such as warfarin, aspirin etc. These patients were instructed to stop taking low molecular weight heparin (LMWH) on the day of blood drawing. The blood should be collected before intestinal clearance, or at three days after colonoscopy (avoid the influence of laxatives and colonoscopy on the blood). After collecting the blood samples (10 ml) using Streck blood collection tubes, these were placed into a sample collection box, and transported to the testing centre (Singlera Genomics, Trademark registration certificate: No. 22604044) at normal temperature (6-37°C). After blood collection, the plasma was separated within 72 hours, the cfDNA in plasma was extracted using a commercial kit (Qiagen, 55114), and the DNA sulfite treatment was performed using another commercial kit (ThermoFisher, MECOV50). Finally, the Illumina platform sequencer was used for sequencing.
Using the data from the tissue and leukocyte reduced representation bisulfite sequencing (RRBS), public database (such as The Cancer Genome Atlas, (TCGA), and methylation markers reported in the literature; 10,613 target differentially methylated genomic regions were obtained by the initial screening, and confirm that the chosen sites truly represented a pan-cancer signature.
Using mTitan patented technology to amplify, the target region of bisulfite treated plasma DNA, and conduct deep sequencing, the average sequencing depth of each target region was found to be above 500X. Then, the methylation haplotype frequency was calculated as a characteristic variable (combinations of methylation states at CpG sites on the same read were usually divided into three haplotypes, respectively, representing the fully methylated haplotypes, non-methylated haplotypes at all sites, and other haplotypes).
The sample was randomly divided into training sets and test sets for the cross-validation. For example, 5X or 10X cross-validation. In each cross-validation, the feature selection was independently performed, and feature variables that have a methylation level consistency in cancer tissues were retained. The machine learning classifier was established using the training set samples, and the test set samples were predicted. Taking the logistic regression model as an example, the selected features were used to construct the regression equation, and the probability threshold was set in order to obtain the best prediction result in the training data. For each test set sample, the regression equation was used to calculate the probability score, and the threshold value obtained before was used to judge the prediction results of the test set sample.
Based on the results of the polygene methylation test in blood and the comprehensive analysis using the existing early screening risk assessment model for CRC, the detected PHF index was calculated, and a value ≥10 was considered positive.
The collected data included the patient’s age, gender, weight, height, hemoglobin level, smoking and drinking history, symptoms (mucous bloody stool, diarrhea and constipation), chronic diseases (hypertension and diabetes mellitus), pathological types, tumor stage, NGS screening results, and FIT and CT examination results. The statistics were analysed using IBM SPSS Statistics version 25.0. The qualitative data was expressed in frequency and percentage, while quantitative data was expressed in median (IQR), along with the mean ± standard deviation (SD). The qualitative data was analysed by Chi-square/Fisher's exact probability test, while quantitative data was analysed by independent sample t-test and Mann-Whitney test. The receiver operating characteristic (ROC) curve was used to evaluate the diagnostic accuracy of NGS for Pre-CRC. The sensitivity, specificity, NPV and PPV of different examinations were also summarized. All statistical tests were bilateral, and p<0.05 was considered statistically significant.
RESULTS
A total of 120 patients from a hospital from June 2019 to February 2020 were included. Among them, 14 (11.7%) participants were included before the surgery of CRC, 13 (10.8%) participants were included after the surgery of CRC, 15 (12.5%) participants had AA, 27 (22.5%) participants had common adenoma, 28 (23.3%) participants had polyps, 12 (10.0%) participants had inflammation, and 11 (9.2%) participants were normal (no lesions under the colonoscopy). The pathological results of the pathological reports were used as the gold standard, and the research revealed that the results of the NGS were statistically different between the preoperative CRC and the normal group, preoperative CRC group and AA group (p=0.002 and 0.011, respectively; Table I). The latter four types (common adenoma, polyp, inflammation and normal group) had a relatively low clinical risk. Hence, these were combined into the control group in the following analysis.
Among the included participants, the youngest participant was of 18 years and the oldest participant was of 86 years, and the average value was 54.41±13.70 years. Seventy-five (62.5%) participants were males, and 45 (37.5%) participants were females. Twenty (16.7%) participants had positive screening results, while 100 (83.3%) participants had negative results. The results revealed that there were no statistically significant differences in basic characteristics, such as age, gender, body mass index (BMI), smoking history and drinking history among the preoperative CRC group, postoperative CRC group, AA group, and control group. The probability of having a history of mucus and bloody stool in the preoperative CRC group and postoperative CRC group was significantly higher, when compared to that in the control group, and the difference was statistically significant. In terms of diarrhea, constipation, diabetes mellitus and coronary heart disease, there were no significant differences among the four groups. In addition, the mean value of hemoglobin in the preoperative CRC group, postoperative CRC group and AA group were lower, when compared to the control group, but the differences were not statistically significant.
The screening results of NGS were statistically different between the preoperative CRC group and the control group, preoperative CRC group and AA group. The screening results of the CT were statistically different in the preoperative CRC group and control group, and the postoperative CRC group and control group. The screening results of the FIT were statistically different only between the preoperative CRC group and control group (Table II).
The screening results of NGS combined with CT were statistically different in the preoperative or postoperative CRC group and control group, preoperative CRC group and AA group, while the results of NGS combined with FIT were statistically different in the preoperative CRC group and control group. The screening results of FIT combined with CT, or the combination of the three methods were both statistically different in the preoperative or postoperative CRC group and control group (Table II).
For the preoperative detection of CRC, the sensitivity of NGS was 57.1%, which was slightly higher than that for CT (45.5%) and FIT (50%). The area under the ROC curve (AUC) was 0.804, the diagnostic value is good, the standard error was 0.077, and the 95% confidence interval was 0.652–0.955 (Figure 1). The sensitivity of NGS combined with CT or FIT could increase to 78.6%. The combined sensitivity of the three methods for preoperative CRC could increase to 92.3%, and the false positive rate (46.4%) also greatly increased (specificity decreased). In addition, NGS and CT both had high specificity for the preoperative detection of CRC (Tables II and III).
For the detection of AA, the sensitivities of NGS, CT and FIT were not high, which were 13.3%, 7.7% and 38.5%, respectively. The sensitivity of NGS combined with CT or FIT was 21.4% and 46.2%, respectively, the sensitivity of FIT combined with CT could reach 71.4%, and the combined sensitivity of the three methods could reach 75.0% (Table II).
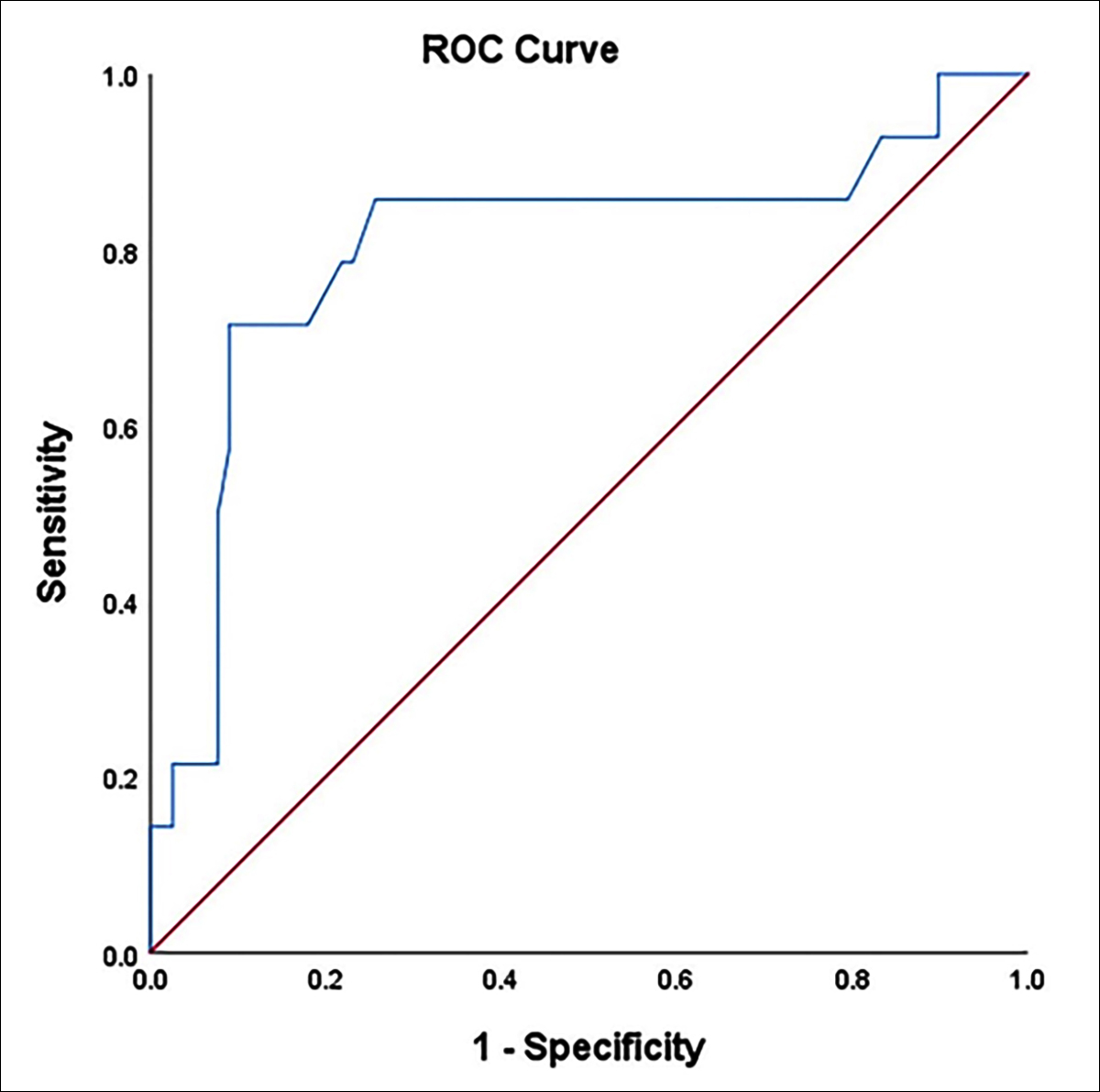 Figure 1: ROC curve for diagnostic performance of NGS for pre-CRC.
Figure 1: ROC curve for diagnostic performance of NGS for pre-CRC.
DISCUSSION
The 5-year relative survival rate for early CRC is 90%, but for advanced CRC, the ratio drops to approximately 14%.13 Therefore, early detection and early diagnosis can significantly improve the prognosis of patients, and are the key factors to reduce the mortality rate of CRC.14 The rapid development of high-throughput sequencing technology provides a technical basis for the comprehensive understanding of the occurrence and development of CRC at the molecular level. Compared with tissue biopsy, liquid biopsy has the advantages of being minimally invasive, simple and fast, overcoming the intratumoral heterogeneity. As the first choice for liquid biopsy, ctDNA is released into the peripheral blood by tumor cell apoptosis, necrosis, or secretion, which carries the genetic variation characteristics of tumor cells.15,16 Previous studies have confirmed that the mutation characteristics of ctDNA in CRC patients is highly consistent with tumor tissue DNA,17 ctDNA detection may replace tissue DNA detection, and play an important role in the precise medical field of CRC. Although ctDNA-based CRC early screening strategy has good application prospects, high detection technology is required due to the low ctDNA content in the early stages of tumors,18 this study would further test its accuracy in the method of NGS.
The results of the present study revealed that NGS has a certain detection capacity for CRC, and the positive detection rate was higher than FIT and CT. Although the positive rate of NGS for AA was only 13.3%, this could reach up to 75.0% after combining with CT and FIT. It can be observed from the results that NGS is complementary to FIT and CT, and the combination of these methods can improve the sensitivity for detecting CRC and AA. The positive rate of NGS in the postoperative CRC group was 23.1%, and this was significantly reduced, when compared to the preoperative CRC group, which may suggest that gene methylation level is significantly correlated with the tumor status in CRC patients, and that NGS can be used to evaluate the efficacy of CRC surgery, or monitor the potential of recurrence and metastasis in CRC patients after treatment. NGS has certain limitations for large-scale screening. However, this provides a new auxiliary detection method for people who are unsuitable or unwilling to receive colonoscopy. Furthermore, with the promotion of clinical application, this may have a high application prospect.
CT examination has the characteristics of fast speed, wide scanning range, and non-invasive. This can clearly show the condition of the lesion and its surrounding organs, and the presence or absence of lymph node metastasis. However, its sensitivity is correlated to the size of the lesion, and the sensitivity for lesion diameters of less than 1 cm is low. Hence, this may be difficult to detect for small tumor lesions of the colorectum or early tumors confined to the cellular level. The results of the present study revealed that the sensitivity of CT for preoperative CRC detection was only 45.5%, but the specificity could reach up to 98.2%, which was higher than NGS and FIT. Therefore, NGS cannot completely replace CT, and that combining these methods may be more suitable for clinical use.
FIT is a popular choice for early CRC screening due to its advantages of simple operation, effectiveness and low cost. However, many false negative results may occur in clinic due to some reasons, such as intermittent hemorrhage of the tumor tissue. FIT often requires repeated tests to improve the sensitivity, which in turn significantly reduces the compliance of patients. In addition, FIT is less sensitive to colorectal tumors which is in the proximal location, and advanced adenomas.19,20 Furthermore, due to the influence of upper gastrointestinal bleeding, other hemorrhagic bowel diseases and hemorrhoid bleeding, FIT may produce many false positive results.21 The results of the present research revealed that the sensitivity of FIT in preoperative CRC detection was 50.0%, and the specificity was 81.8%, and both of which were lower than NGS. FIT alone cannot achieve the goal of early CRC screening. However, after the combination of these three, the sensitivity could reach up to 92.3%. Therefore, it is suggested that these detection methods should be used in combination with each other, according to the patient's condition and economic status, rather than replacing each other.
There were some limitations in the present study. First, the present study had a small sample size. Furthermore, the participants come from a hospital, which may cause certain bias, regardless of whether the result is consistent with the general risk population, requiring further in-depth studies for verification.
CONCLUSION
NGS can improve the diagnostic accuracy of CRC, and CT or FIT alone cannot achieve the goal of early CRC screening. The sensitivity can reach up to 92.3% after the combination of the three methods was used. For AA, the accuracy of NGS was limited. Hence, this should be diagnosed in combination with other items. Can NGS replace FIT or CT in the screening of colorectal cancer and advanced adenoma? Further studies are needed to determine this. The results suggest that the detection methods should be used in combination with each other, according to the patient's condition and economic status, rather than replacing each other.
ETHICAL APPROVAL:
The study was approved by the Ethics Committee of the Renmin Hospital of Wuhan University prior to initiation of the research work. All procedures performed in the present study, which involved human participants, were conducted in accordance to the ethical standards of the Institutional Research Committee, and with the 1964 Helsinki Declaration and its later amendments.
PATIENTS' CONSENT:
All patients provided a signed informed consent, prior to testing, and agreed to publish the data.
CONFLICT OF INTEREST:
The authors report no conflict of interest.
AUTHORS' CONTRIBUTION:
XLY: Acquired and analysed data; drafted manuscript and approved the final manuscript.
HC: Acquired data; approved the final manuscript.
CYH, ML: Analysed data; approved the final manuscript.
TD: Drafted manuscript; read and approved the final manu-script.
REFERENCES
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65(2):87-108. doi: 10.3322/caac.21262.
- Ellis L, Abrahao R, McKinley M, Yang J, Somsouk M, Marchand LL, et al. Colorectal cancer incidence trends by age, stage, and racial/ethnic group in california, 1990-2014. Cancer Epidemiol Biomarkers Prev 2018; 27(9):1011-8. doi: 10.1158/1055-9965.EPI-18-0030.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68(6):394-424. doi: 10. 3322/caac.21492.
- Brenner H, Hoffmeister M, Stegmaier C, Brenner G, Altenhofen L, Haug U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: Estimates based on 840,149 screening colonoscopies. Gut 2007; 56(11): 1585-1589. doi: 10.1136/gut.2007.122739.
- Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, et al. Colorectal Cancer Screening: Recommendations for Physicians and Patients from the U.S. Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol 2017; 112(7):1016-1030. doi: 10.1038/ajg. 2017.174.
- Xie L, Jiang X, Li Q, Sun Z, Quan W, Duan Y, et al. Diagnostic value of methylated septin9 for colorectal cancer detection. Front Oncol 2018; 8:247. doi: 10.3389/ fonc.2018.00247.
- Wild N, Andres H, Rollinger W, Krause F, Dilba P, Tacke M, et al. A combination of serum markers for the early detection of colorectal cancer. Clin Cancer Res 2010; 16(24): 6111-21. doi: 10.1158/1078-0432.CCR-10-0119.
- Simons CC, Hughes LA, Smits KM, Khalid-de Bakker CA, de Bruine AP, Carvalho B, et al. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: Implications for prognosis. Ann Oncol 2013; 24(8):2048-56. doi: 10.1093/annonc/mdt076.
- Dong L, Ren H. Blood-based DNA Methylation biomarkers for early detection of colorectal cancer. J Proteomics Bioinform 2018; 11(6):120-6. doi: 10.4172/jpb.1000477.
- Sun J, Fei F, Zhang M, Li Y, Zhang X, Zhu S, et al. The role of (m) SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 2019; 19(1):450. doi: 10.1186/s12885-019-5663-8.
- Hashimoto Y, Zumwalt TJ, Goel A. DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics 2016; 8(5):685-703. doi: 10.2217/epi-2015-0013.
- Yokoi K, Yamashita K, Watanabe M. Analysis of DNA methylation status in bodily fluids for early detection of cancer. Int J Mol Sci 2017; 18(4). doi: 10.3390/ijms 18040735.
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017; 67(3):177-93. doi: 10.3322/caac.21395.
- Moore JS, Aulet TH. Colorectal cancer screening. Surg Clin North Am 2017; 97:487-502.
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17(4):223-38. doi: 10.1038/nrc.2017.7
- Yoruker EE, Holdenrieder S, Gezer U. Blood-based biomarkers for diagnosis, prognosis and treatment of colorectal cancer. Clin Chim Acta 2016; 455:26-32. doi: 10.1016/j.cca.2016.01.016.
- Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012; 18(4):1177-85. doi: 10.1158/1078- 0432.CCR-11-0564.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6(224):224ra224. doi: 10.1126/scitranslmed. 3007094.
- Kim NH, Yang HJ, Park SK, Park JH, Park DI, Sohn CI, et al. Does Low Threshold Value Use Improve Proximal Neoplasia Detection by Fecal Immunochemical Test? Dig Dis Sci 2016; 61(9):2685-93. doi: 10.1007/s10620-016-4169-3.
- Castro I, Cubiella J, Rivera C, Gonzalez-Mao C, Vega P, Soto S, et al. Fecal immunochemical test accuracy in familial risk colorectal cancer screening. Int J Cancer 2014; 134(2): 367-75. doi: 10.1002/ijc.28353.
- Van Turenhout ST, Oort FA, Terhaar sive Droste JS, Coupe VM, van der Hulst RW, Loffeld RJ, et al. Hemorrhoids detected at colonoscopy: An infrequent cause of false-positive fecal immunochemical test results. Gastrointest Endosc 2012; 76(1):136-43. doi: 10.1016/j.gie.2012. 03.169.